Import annotations from NIfTI files
A CLI how-to guide. SDK power-users can refer to our full SDK docs generated from our source code here
Ensure your images and videos have been successfully uploaded and processedBefore you import annotations, make sure your target images and videos are marked as
New, or have been previously annotated already.
Once you've added volumetric data to a dataset in V7, you can use CLI commands to import existing annotations from NIfTI files. You'll need to create a .json file containing:
- The paths to the local NifTI volume annotations
- The paths to the remote dataset files to import the annotations to
- A class mapping that maps integer values to Darwin class names
- A mode to import annotations as either:
video- Each class will result in a single continuous polygon annotation. This makes sense for contiguous 3D objects like a whole organ or a single tumorinstances- Each class instance will result in a single polygon annotation. This makes sense for scenarios where the same class may appear many timesmask- Each class will result in a mask annotation written to the raster layer. For more information on masks please see our article on masks
Here is an example of what such a .json file should look like:
{
"data": [
{
"image": "/remote/path/to/dataset/item/volume.nii",
"label": "/local/path/to/nifti/annotations/annotations.nii.gz",
"slot_names":["name_of_target_slot"],
"class_map": {
"0": "background",
"1": "oedema",
"2": "non-enhancing tumor",
"3": "enhancing tumor",
},
"mode": "video"
}
]
}It's important to note:
- The
imagepath needs to match the remote filepath used in the Darwin dataset exactly - The
labelpath is a local relative path from where you are running thedarwin dataset importcommand slot_namesdetermines the slot that the NifTI annotation will be imported to. If not provided, the annotations will be uploaded to the first medical slot in the dataset item- The
backgroundclass is not imported as annotations
Multiple NIfTI uploads at onceYou can add NIfTI labels for multiple files in a dataset with a single call to
darwin dataset import, just add more entries in thedatalist with more paths and class maps.
You can now run the only CLI command needed to upload
darwin dataset import your-team/your-dataset nifti upload.jsonIf the class names provided do not match those created in your team, the CLI will prompt you to create the missing classes.
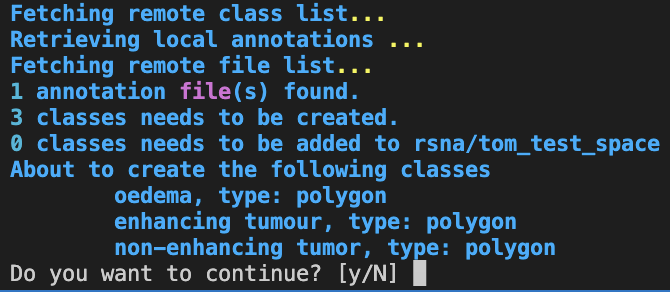
That’s it! You should now be able to view your 3D annotations on the platform.
Updated 5 months ago
